Archive
How to install Greg Mori’s superpixel MATLAB code?
This short note aims to show you how to use superpixel code from Greg Mori whose codes are observed to have very good results and used by a bunch of computer vision researchers. However, the installation process can be challenging sometimes ^_^, so I figured it’d be nice if I document the process so that it will be easier for absolute beginners to use the code, and more importantly…I can come back to read when I forget how to do it.
I have MATLAB R2010a installed on my Ubuntu 32-bit 10.04 LTS – the Lucid Lynx
I download Mori’s code, extract the zipped file to a folder called superpixels. The folder is located at
/home/student1/MATLABcodes/superpixels
Next I download the boundary detector code from the link
http://www.eecs.berkeley.edu/Research/Projects/CS/vision/grouping/segbench/code/segbench.tar.gz
I extract it to a folder called segbench, then I put it inside the superpixels
/home/student1/MATLABcodes/superpixels/segbench
—————————————————————————–
Here are the instructions of the folders
README (Mori’s)
– Run mex on *.c in yu_imncut directory
– Obtain mfm-pb boundary detector code from
http://www.cs.berkeley.edu/projects/vision/grouping/segbench/
– Change path names in sp_demo.m and pbWrapper.m
– Get a fast processor and lots of RAM
– Run sp_demo.m
README (segbench’s)
(1) For the image and segmentation reading routines in the Dataset
directory to work, make sure you edit Dataset/bsdsRoot.m to point to
your local copy of the BSDS dataset.
(2) Run ‘gmake install’ from this directory to build everything. You
should then probably put the lib/matlab directory in your MATLAB path.
(3) Read the Benchmark/README file.
———————————————————————————————————————
According to the README instruction
– Run mex on *.c in yu_imncut directory
I run mex on all the .c file in the folder
/home/student1/MATLABcodes/superpixels/yu_imncut
I don’t know why the command mex *.c does not work, so I have to run mex on every file one by one. Each time I run mex, I will get message
“Warning: You are using gcc version “4.4.3-4ubuntu5)”. The version
currently supported with MEX is “4.2.3”.
For a list of currently supported compilers see:
http://www.mathworks.com/support/compilers/current_release/”
However, it seems to work fine since I can see all the .mexglx files show up in the folder. So I assume I do it correctly and go on the next step.
In this step,
– Obtain mfm-pb boundary detector code from
http://www.cs.berkeley.edu/projects/vision/grouping/segbench/
I got the code already, so I follow the README (segbench’s). Firstly, I do
(1) For the image and segmentation reading routines in the Dataset
directory to work, make sure you edit Dataset/bsdsRoot.m to point to
your local copy of the BSDS dataset.
So, I go to the file /home/student1/MATLABcodes/superpixels/segbench/Dataset/bsdsRoot.m and change the root to
root = ‘/home/student1/MATLABcodes/superpixels’;
which contains the image I want to segment, “img_000070.jpg”
Next, I do (2) in README (segbench’s)
(2) Run ‘gmake install’ from this directory to build everything. You
should then probably put the lib/matlab directory in your MATLAB path.
Now at the folder, student1@student1-desktop:~/MATLABcodes/superpixels/segbench$
we need to make MATLAB seen in this folder, so we export the MATLAB path
student1@student1-desktop:~/MATLABcodes/superpixels/segbench$ PATH=$PATH:/usr/share/matlabr2010a/bin
student1@student1-desktop:~/MATLABcodes/superpixels/segbench$ export PATH
then use make install, this time I got quite a long message in the terminal
student1@student1-desktop:~/MATLABcodes/superpixels/segbench$ make install
Then you will notice some files in the folder
/home/student1/MATLABcodes/superpixels/segbench/lib/matlab
What you have to do here is to addpath in MATLAB by typing in the command window
addpath(’/home/student1/MATLABcodes/superpixels/segbench/lib/matlab’);
Next, (3) Read the Benchmark/README file. I found that we don’t have to do anything in this step. So just skip this.
Now it’s the last step
Change path names in sp_demo.m and pbWrapper.m
so, go to the folder /home/student1/MATLABcodes/superpixels and change the path
in pbWrapper.m I make the path pointing to ‘/home/student1/MATLABcodes/superpixels/segbench/lib/matlab’
in sp_demo.m I make the path pointing to
‘/home/student1/MATLABcodes/superpixels/yu_imncut’
Now run the file sp_demo.m. Unfortunately you will get some error messages because of a function spmd. This happens because MATLAB 2010a has function spmd of its own which has the number of input argument different from that of spmd from the toolbox. One way to get around this is to change the name of spmd.c in the toolbox to spmd2.c, then compile spmd2.c using mex spmd2.c. Then replace spmd(…) with spmd2(…). If you encounter more errors from this point on, don’t panic, because it’s probably from this spmd issue, so just do the same thing and it will work fine.
That’s it! Enjoy Greg Mori’s code!
——————————————————————————
For windows users, please refer to Thanapong’s blog, whose URL is given below:
http://blog.thanapong.in.th/arah/?p=29
Image Segmentation using Gaussian Mixture Model (GMM) and BIC
A while ago, I was so amazed about the image segmentation results using Gaussian Mixture Models (GMMs) because GMM gives pretty good results on normal/natural images. There are some results on my previous post. Of course, GMM is not the best for this job, but hey look at its speed and easiness to implement–it’s pretty good in that sense. However, one problem with GMM is that we need to pick the number of components. In general, the more component numbers we assume, the better log-likelihood it would be for GMM. In that case, we would simply send the number of components to infinity, right? Well…but there is nothing good come out of that because the segment would not be so meaningful–in fact, we overfit the data, which is bad.
Therefore, Bayesian Information Criteria (BIC) is introduced as a cost function composing of 2 terms; 1) minus of log-likelihood and 2) model complexity. Please see my old post. You will see that BIC prefers model that gives good result while the complexity remains small. In other words, the model whose BIC is smallest is the winner. Simple as that. Here is the MATLAB code. Below are some results from sweeping the number of components from 2 to 10. Unfortunately, the results are not what I (and maybe other audiences) desire or expect. As a human, my attention just focuses on skier, snow, sky/cloud and perhaps in the worst case, the shadows, so the suitable number of components should be 3-4. Instead, the BIC assigns 9-component model the winner which is far from I expected. So, Can I say that the straightforward BIC might not be a good model for image segmentation, in particular, for human perception? Well…give GMM-BIC a break– I think this is too early to blame BIC because I haven’t use other more sophisticated features like texture, shape, color histogram which might improve results from using GMM-BIC. The question is what are the suitable features and the number of components that makes the segmentation results using GMM-BIC similar to human perception? MATLAB code is made available here.

original image

Plot of BIC of model using 7-10 components
Irregular Tree Structure Bayesian Networks (ITSBNs)
Irregular Tree Structure Bayesian Networks (ITSBNs)
This is my on-going work on structured image segmentation. I’m about to publish the algorithm soon, so the details will be posted after submission. ^_^ Please wait.
<details will be appeared soon>
Here are some results

original image

segmented using GMM

segmented using ITSBN
How to use the ITSBN toolbox
- Install Bayesian networks MATLAB toolbox and VLFeat. Let’s say we put them in the folders Z:\research\FullBNT-1.0.4 and Z:\research\vlfeat-0.9.9 respectively.
- Download and unpack the ITSBN toolbox. Let’s say the folder location is “Z:\research\ITSBN”. The folder contains some MATLAB files and 2 subdirectories 1) Gaussian2Dplot and 2) QuickShift_ImageSegmentation
- Put any color image to be segmented in the same folder. In this case, we use the one from Berkeley image segmentation BSDS500 and the folder is ‘Z:\research\BSR\BSDS500\data\images\test’
- Open the file main_ITSBNImageSegm.m in MATLAB and make sure that all the paths pointing to their corresponding folders:
- vlfeat_dir = ‘Z:\research\vlfeat-0.9.9\toolbox/vl_setup’;
- BNT_dir = ‘Z:\research\FullBNT-1.0.4’;
- image_dataset_dir = ‘Z:\research\BSR\BSDS500\data\images\test’;
- Run main_ITSBNImageSegm.m. When finished you should see folders of segmented images in the folder ‘Z:\research\ITSBN’.
Often-used MATLAB Commands
I’m very forgetful…Hopefully this will help me to fetch some memory when I want to use it.
mkdir -- make a directory
../testdata -- go out one step
save([imagename,'_package'], 'Y', 'Y_index', 'Z', 'evidence_label', 'n_level', 'H');
movefile(['./',imagename,'_package.mat'], ['./',imagename]);
copyfile('source','destination')
[pathstr, name, ext] = fileparts(filename)
[s,mess,messid] = mkdir('../testdata','newFolder')
pwd -- identify current folder
list all the filename with extension from a directory and its subdirectory. Please visit [link] from stackoverflow.
caxis([cmin cmax])
How do I control axis tick labels, limits, and axes tick locations?
http://www.mathworks.com/support/solutions/en/data/1-15HXQ/index.html
move xtick label to the top
set(gca,'XAxisLocation','top');
How to plot image icon on the image? I will post my code very soon.
Making Superpixels for An Image
Nowadays it seems to me that people in computer vision community, especially who work on image segmentation, analysis, interpreting and classification, tend to adopt the idea of using superpixels rather than using raw pixels. That because superpixels can significantly reduce computational load of an algorithm, and are cheap to produce. In my work, image segmentation using graphical model, superpixels can potentially save me a lot of time running inference/learning algorithm tremendously. There are off-the-shelf superpixel algorithms available on the internet. I’m using QuickShift from VLFEAT toolbox and it works fine after manually tuning a couple parameters. Here are some results.

Segmentation result overlaid on the original image.
However, it is hard to find when you are in need…Therefore, I figure that it is a good idea to put those algorithms together in this post. To be honest with you, I haven’t been using them lately, so I forgot a lot. Well, I plan to add up and grow the collection on day by day basis. So, you are very welcome to suggest any algorithm you like. Let’s make this collection together!
Pablo Arbelaez’s UCM [link] — I have a problem installing it on my machine…
QuickShift in VLFEAT [link] — very convenient MATLAB toolbox
Segmentation by Minimum Code Length [link]
Greg Mori’s superpixel code [link]
Turbopixel [link]
Scale-Invariant Image Representation: the CDT Graph [link]
There are also some empirical studies on superpixel talking about how many superpixels should be in an image, advantages, disadvantages, behavior, etc., available. Here are my favorites:
Superpixel: Empirical Studies and Applications [link]
On Parameter Learning in CRF-based Approaches to Object Class Image Segmentation [link] by Nowozin and Lampert
and chapter4 of the (draft) book “Structured Learning and Prediction in Computer Vision” [link] by Nowozin and Lampert.
Balanced-Tree Structure Bayesian Networks (TSBNs) for Image Segmentation
In this work, I made a more generalized version of Gaussian mixture model (GMM) by putting prior balanced-tree structure over the class variables (mixture components) in the hope that the induced correlation among the hidden variables would suppress the noise in the resulting segmentation. Unlike supervised image classification by [1], this work focuses on totally unsupervised segmentation using TSBN. In this work, it is interesting to see how the data will be “self-organized” according to the initial structure given by TSBN.
The MATLAB code is available here. The codes call inference routines in Bayesian network toolbox (BNT), so you may want to install the toolbox before using my TSBN code.

original image 136x136

16x16 feature image. Each pixel represents 16-dimensional vector.

Segmentation result using TSBN by setting predefined number of classes to 3. It turns out that the classes are meaningful as they are sky, skier and snow.
[1] X. Feng, C.K.I. Williams, S.N. Felderhof, “Combining Belief Networks and Neural Networks for Scene Segmentation,” IEEE Transactions on Pattern Analysis and Machine Intelligence, pp. 467-483, April, 2002
fit Gaussian mixture Models (GMMs) with known initial parameters using MATLAB
Unfortunately the description on using this feature is not so clear in MATLAB help. I found this link very helpful.
http://www.mathworks.com/matlabcentral/newsreader/view_thread/244946
I also make an easy example whose the results are shown below. The MATLAB code is available here. In the first figure, I fit gmm to the data point Y. Next, I add some new points (red) to the data point Y, let’s call the new data point Y_new. In second figure, Y_new is fitted with gmm with known initial parameters obtained from the previous experiment. In the last figure, Y_new is fitted with gmm with random initial parameters.
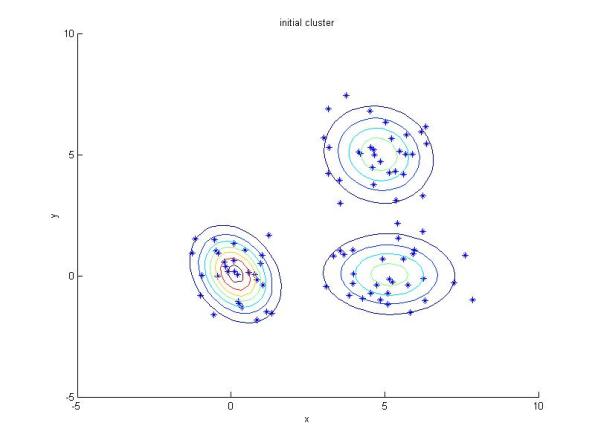
Resulting clusters using GMM with random intial parameters.
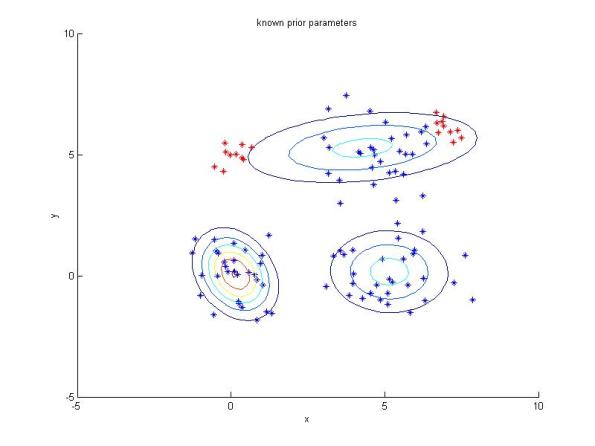
Resulting clusters for the newe dataset with new points (red) added using GMM with initial parameters obtained from the previous result.

Resulting clusters for the newe dataset with new points (red) added using GMM with random initial parameters.
3D LIDAR Point-cloud Segmentation
One of the big challenges in 3D LIDAR point-cloud segmentation is detailed ground extraction, especially in high vegetated area. In some applications, it requires to extract the ground points from the LIDAR data such that the details are preserved as much as possible, however, most of the time the details and the noise are coupled and it is difficult to remove the noise whereas the ground details are preserved. Imagine the case where you have the LIDAR point cloud over a creek covered by multilayer canopies including ground flora and you would like to extract the creek from the data set by preserving the ground details as much as you can. This would be a very labor-intensive task for human, so a better choice might be to develop an automatic process for computer to complete the task for us. Even for a computer, this can be a very labor-intensive task due to the number of points in the area is extremely high.


In 2004, I and my former adviser, Dr. Kenneth C. Slatton, developed a multiscale information-theoretic based algorithm for ground segmentation. The method works well in real-world applications and is used in several publications. The MATLAB toolbox is available here. The brief manual can be found here.
I would like to thank my colleagues at National Center for Airborne Laser Mapping (NCALM), Adaptive Signal Processing Laboratory (ASPL) and Geosensing group at University of Florida who use the algorithm on their work and give tons of useful suggestions to improve this algorithm up until now; Dr. Jhon Caceres for very nice GUI; Dr. Sowmya Selvarajan for the first-ever manual for this toolbox. Last but not least, I would like to thank Dr. Kenneth Clint Slatton for wonderful ideas and guidance–we still have an unpublished journal to fulfill [1].
/* Style Definitions */
table.MsoNormalTable
{mso-style-name:”Table Normal”;
mso-tstyle-rowband-size:0;
mso-tstyle-colband-size:0;
mso-style-noshow:yes;
mso-style-priority:99;
mso-style-qformat:yes;
mso-style-parent:””;
mso-padding-alt:0in 5.4pt 0in 5.4pt;
mso-para-margin-top:0in;
mso-para-margin-right:0in;
mso-para-margin-bottom:10.0pt;
mso-para-margin-left:0in;
line-height:115%;
mso-pagination:widow-orphan;
font-size:11.0pt;
mso-bidi-font-size:14.0pt;
font-family:”Calibri”,”sans-serif”;
mso-ascii-font-family:Calibri;
mso-ascii-theme-font:minor-latin;
mso-fareast-font-family:”Times New Roman”;
mso-fareast-theme-font:minor-fareast;
mso-hansi-font-family:Calibri;
mso-hansi-theme-font:minor-latin;}
[1] K. Kampa and K. Clint Slatton, “Information-Theoretic Hierarchical Segmentation of Airborne Laser Swath Mapping Data,” IEEE Transactions in Geoscience and Remote Sensing, (in preparation).
[2] K. Kampa and K. C. Slatton, “An Adaptive Multiscale Filter for Segmenting Vegetation in ALSM Data,” Proc. IEEE International Geoscience and Remote Sensing Symposium (IGARSS), vol. 6, Sep. 2004, pp. 3837 – 3840.
A brief slides can be found here.
Bayesian Inference
Recently I have been looking for a good short resource for the fundamental Bayesian inference. There are tons of good relevant materials online, unfortunately, some are too long, some are too theoretic. Come on, I want to find something intuitive, making sense and understandable and readable within 30 minutes! After a couple of hours wandering on internet and reading some good relevant books, I decided to pick the best 3 books, of course in my opinion, that contributes to my understanding the most within half an hour.
- Bayesian Data Analysis by Andrew Gelman et al. — If you understand the concept of Bayesian framework and just want to review Bayesian inference, then the first pages of Chapter2 are sufficient. Good examples can be found in section 2.6.
- Bayesian core: a practical approach to computational Bayesian statistics by Jean-Michel Marin, Christian P. Robert — The book emphasizes the important concepts and has a lot of examples necessary to understand the basic ideas.
- Bayesian Field Theory by Jörg C. Lemm — This book provides great perspective of Bayesian framework to learning, information theory and physics. There are very a lot of figures and graphical models making the book easy to follow.
GMM-BIC: A simple way to determine the number of Gaussian components
Gaussian Mixture Models (GMMs) are very popular in broad area of applications because its performance and its simplicity. However, it is still an open problem on how to determine the number of Gaussian components in a GMM. One simple solution to this problem is to use Bayesian Information Criteria (BIC) to penalize the complexity of the GMM. That is, the cost function of BIC-GMM is composed of 2 parts: 1) log-likelihood and 2) complexity penalty term. Consequently, the final GMM would be a model that can fit the data well, but not “overfitting” the model in BIC sense. There are tons of tutorials on the internet. Here I would like to share my MATLAB code for demo.
Note that Variational Bayes GMM (VBGMM) can also solve this problem in a different flavor and is worth to study and compare with GMM-BIC. I also provided some details of the derivations of VBGMM here.

















